19 - Longer negative feedback circuits#
We saw in the previous section that a two-component negative feedback circuit (the auto-repressive gene system) possesses a unique steady state, which is always stable. This underlies homeostasis; the tendency of a negative feedback system to return to its “set point” when perturbed (for small perturbations, at least).
However, negative feedback circuits can also generate sustained oscillations. One way this can occur is if we extend our two-variable model to a model including at least three variables. This was first proposed in the context of regulated gene expression by the theoretical biologist Brian Goodwin in the 1960s, and the prototype three-component model is often referred to as the Goodwin oscillator. There are a number of ways of thinking about what might underlie a model of auto-regulatory gene expression that would contain more than two variables. One example would be that in fact the protein product of the gene is translated/produced in one place - the cytoplasm - but regulates transcription in another - the nucleus. We could therefore consider three variables: mRNA, cytoplasmic protein and nuclear protein.
A simplified 3-variable model#
Here, we will look at a slightly different model to Goodwin’s original, but the principles remain the same. Rather than focusing on a specific model of gene expression, we will consider a simple model of three interacting components, \(X\), \(Y\) , and \(Z\), which form a negative feedback loop by regulating each other. In particular:
Increasing \(X\) leads to increased production of \(Y\),
Increasing \(Y\) leads to increased production of \(Z\),
Increasing \(Z\) leads to decreased production of \(X\).
Put together there is therefore a negative feedback in the system (since increasing \(X\) ultimately leads to decreased production of \(X\)). For simplicity we will assume that the degradation rates of all three components are the same, so our model can be written as,
Each of the functions \(f_i\) are non-negative and bounded as we assumed before. Given our assumptions in the bullet points we also have that \(df_1/dZ<0\), \(df_2/dX>0\) and \(df_3/dY>0\).
Equilibria of this system occur where the following three conditions are simultaneously satisfied:
How many equilibria can we expect? If we let \(F_i=f_i/\mu\), then we have \(X=F_1(F_3(F_2(X)))=G(X)\). We know that \(G(X)\) must be a decreasing function of \(X\) (intuitively it must be because we know we have a negative feedback loop, but we can also show it by the chain rule). By the same argument as in the two-component auto-repression model, then, we can say that there must be a single equilibrium (because \(X\) is an increasing straight line and \(G(X)\) is some decreasing function, they can only cross once).
Linear stability analysis#
The Jacobian for this system may be 3 dimensional but doesn’t look too horrid,
where \(\phi_i\) are the derivatives of \(f_i\) at the equilibrium point. We have seen before that 3-dimensional Jacobians cannot be solved as easily with trace and determinant conditions (reminder: if you want to look into this more, look up the Routh-Hurwitz criteria). However, because we kept this system relatively simple we can actually solve it from first principles, as it were. To do this we find the characteristic equation of the Jacobian,,
We then look for solutions for \(\lambda\) to where this equation equals 0. That is,
Here we have made the product of our three gradients equal to a new parameter \(-\chi^3\). The minus sign is to remind us that this product is negative (so in fact \(\chi>0\)) and the cubed is to remind us that this is really the composite of three parameters. The solution to this is then,
Somewhat susprisingly then, we find ourselves needing to find the cubed root of -1 in order to understand a real-world biological problem. Recalling that we can write \(e^{-i\pi}=-1\) (I bet you didn’t expect to see Euler’s relation during this course!), we have that the three possible solutions are,
\(\lambda_1 = -\chi-\mu\),
\(\lambda_2 = e^{i\pi/3}\chi-\mu\),
\(\lambda_3 = e^{-i\pi/3}\chi-\mu\).
We can see that \(\lambda_1\) is definitley negative. However the other two eigenvalues are complex, and we can find that they have real part \(Re(\lambda_{2,3})=-\mu+\chi/2\). Putting this all together we find that,
if \(\chi<2\mu\), all the eigenvalues have negative real part and the equilibrium is a stable spiral (we know it is a sprial because two of the eigenvalues are complex),
if \(\chi>2\mu\), two of the eigenvalues have positive real part and the equilibrium is an unstable spiral.
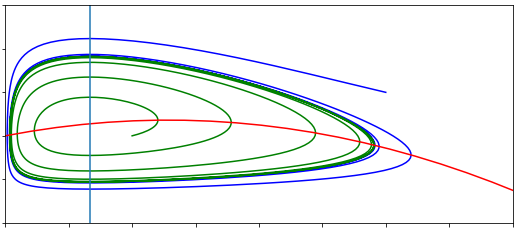
We have seen a case before where stability moves from a stable spiral to an unstable spiral, when we looked at the predator-prey model. You may recall this produced a Hopf bifurcation, where a limit cycle emerges. The same phenomenon occurs here, with sustained oscillations in the expression of the gene arising.
The three-component feedback loop we have considered here demonstrates that negative feedbacks can do more than homeostasis (stability of a system around a set point). It can also result in sustained oscillations. Such oscillations are a central feature of many biological systems. A prominent example is the roughly 24-hour period circadian oscillatory gene expression that occurs in both plants and animals. These oscillations, which are critical for coordinating metabolic activity, etc. with the rotation of the Earth, are generated by a slight elaboration of the mechanism we have studied here. Indeed, the Goodwin oscillator provided an early model for studying the properties of circadian oscillations. Other oscillations, with a wide range of oscillatory periods, are also generated by negative feedbacks.
Have a go: coding
To use the code, click the rocket icon at the top of the page, and then click Colab. This will take you to a live version of this page where you can run the code. Scroll down to the code cell below and either click in the cell and hit Shift-Enter, or click on the ‘Play’ icon in the top-left of the cell. You may see a warning about loading content from Github - click ‘Run anyway’.
This code will produce a time-course and phase portrait for the dynamics of a 3 variable genetic circuit with a negative feedback. The dynamics are governed by the following ordinary differential equations:
You will see a set of sliders to choose the values of the parameters, and boxes for you to enter initial conditions. When you have chosen your values you will see two plots appear - a time course for the three variables, and a phase portrait (in the space of \(X\) and \(Y\). Starting with the given value of \(n=1\), slowly increase \(n\), making the regulation functions steeper. You should see the limit cycles emerge as you move to higher values of \(n\).
Show code cell source
#@title
# Import the necessary libraries
import numpy as np
import matplotlib.pyplot as plt
from scipy.integrate import odeint
import ipywidgets as widgets
from ipywidgets import interactive
muw=widgets.FloatSlider(min=0.5, max=1.5, step=0.1, value=1, description='mu')
thetaw=widgets.FloatSlider(min=0.01, max=0.2, step=0.01, value=0.1, description ='theta')
nw=widgets.FloatSlider(min=1, max=5, step=0.5, value=1,description='n')
X0_w=widgets.BoundedFloatText(value=0,min=0,max=2,description='X0)')
Y0_w=widgets.BoundedFloatText(value=0,min=0,max=2,description='Y0')
Z0_w=widgets.BoundedFloatText(value=0,min=0,max=2,description='Z0')
u1 = widgets.HBox([X0_w, Y0_w, Z0_w])
#Lotka-Volterra dynamics
def goodwin(x,t,mu,theta,n):
X=x[0]
Y=x[1]
Z=x[2]
dX = -mu*X + theta**n/(theta**n+Z**n)
dY = -mu*Y + X**n/(theta**n+X**n)
dZ = -mu*Z + Y**n/(theta**n+Y**n)
return [dX,dY,dZ]
def rungoodwin(mu,theta,n,X0,Y0,Z0):
N0=[X0,Y0,Z0]
tc = np.linspace(0, 50, 1000)
Nc = odeint(goodwin, N0, tc,args=(mu,theta,n))
plt.rcParams['figure.figsize'] = [12, 4]#
plt.rcParams.update({'font.size': 16})
fig, (ax1, ax2) = plt.subplots(1, 2)
ax1.plot(tc, Nc[:,0], "r", label="X")
ax1.plot(tc, Nc[:,1], "k", label="Y")
ax1.plot(tc, Nc[:,2], "b", label="Z")
ax1.set(xlabel='Time', ylabel='Concentrations')
ax1.legend()
ax1.axis([0,50,0,1])
ax2.plot(Nc[:,0],Nc[:,1],'b')
ax2.axis([0, 2, 0, 2])
ax2.set(xlabel='X', ylabel='Y')
ax2.axis([0,1,0,1])
plt.show()
return()
out = widgets.interactive_output(rungoodwin, {'mu': muw, 'theta': thetaw, 'n': nw, 'X0': X0_w, 'Y0': Y0_w, 'Z0': Z0_w})
display(nw)
display(muw,thetaw)
display(u1, out)
3 key points#
We can create a 3 vairable negative feedback circuit with two positive regulatory functions and one negative.
In a simplified system we can calculate the eigenvalues by finding solutions to the characteristic equation of the Jacobian.
We find limit cycles can emerge for steep enough regulation funcions, such that gene expression regularly oscillates.